Discover Cytoscape: Network Visualization & Analysis Platform | Guide
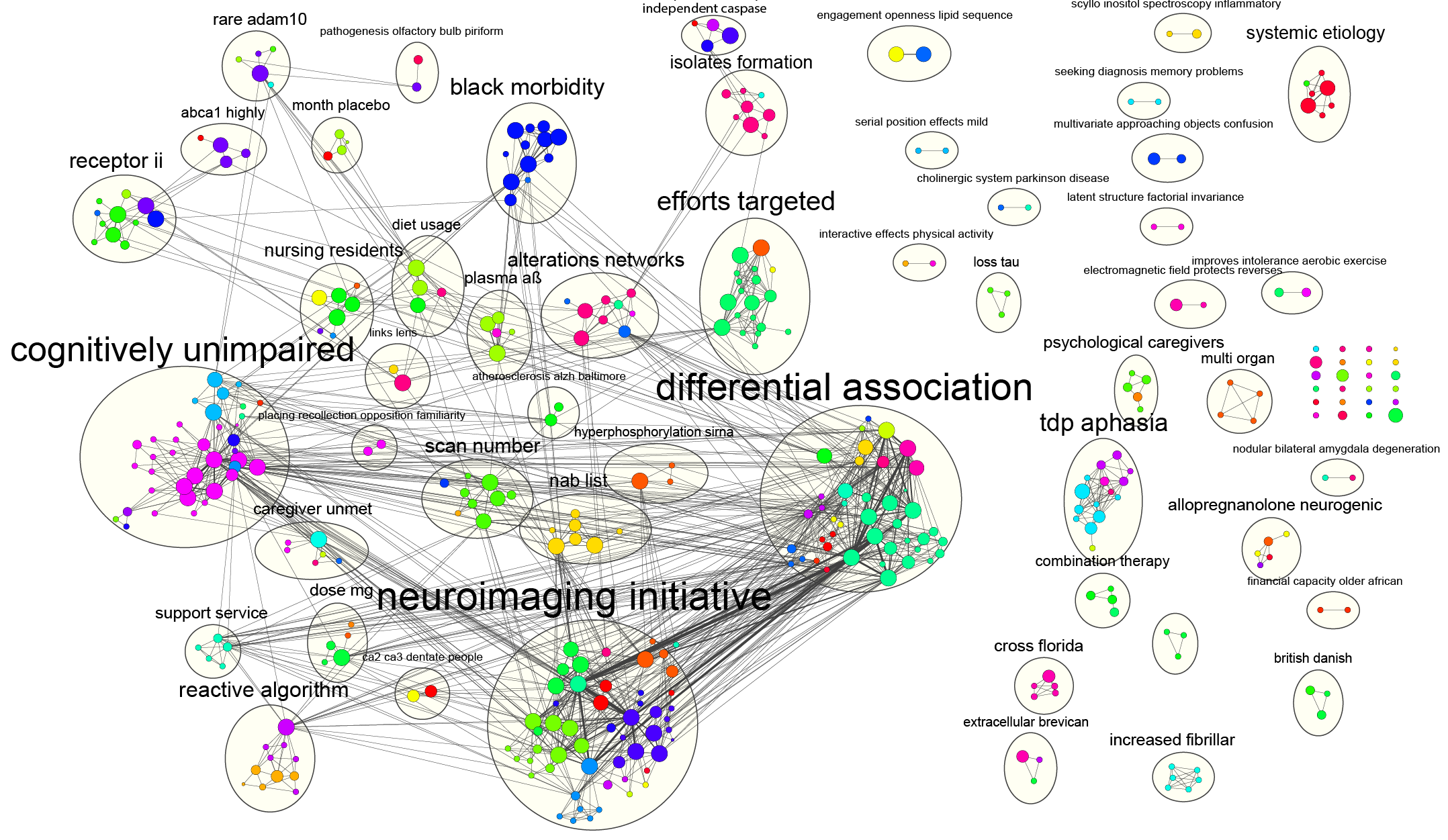
Is there a single tool capable of unraveling the intricate web of complex networks, from the microscopic realm of biological interactions to the vast expanse of social connections? Cytoscape emerges as a powerful and versatile open-source platform designed to visualize and analyze these complex networks, offering researchers and analysts a unique window into the interconnectedness of data.
Initially conceived for biological research, Cytoscape has evolved into a versatile tool, now serving as a general platform for complex network analysis and visualization across diverse fields. Its core distribution provides a foundational set of features for data integration, enabling users to seamlessly import, manipulate, and explore complex datasets. With a broad spectrum of applications spanning bioinformatics, social network analysis, and semantic web explorations, Cytoscape offers a multifaceted approach to understanding intricate relationships within data.
| Feature | Details |
|---|---|
| Software Name | Cytoscape |
| Type | Open Source Software Platform |
| Primary Function | Visualization and analysis of complex networks |
| Original Design | Biological Research |
| Current Use Cases | Bioinformatics, Social Network Analysis, Semantic Web, and more. |
| Key Features | Data integration, network visualization, attribute data analysis, and app extensibility. |
| Developer | International Consortium of Open Source Developers |
| License | GPL License |
| Latest Version at Time of Article | Cytoscape 3.0 (introduces a new architecture) |
| User Manual Availability | Cytoscape 3.10.3 and 3.9.1 user manuals are available |
| Core Capabilities | Basic features for data integration. |
| Network Creation Methods | Importing data from databases, creating empty networks manually. |
| Supported Platforms | Linux, Windows, macOS |
| Additional Platform Compatibility | May run on other UNIX platforms (e.g., Solaris, FreeBSD) with Java version 8. |
| First Appearance | Institute of Systems Biology, Seattle, 2002 |
| App Ecosystem | Extensive apps available for various problem domains |
| User Controls | Provides a set of user controls to perform various task |
| Tutorials and Guides | Quick tour of cytoscape, basic expression analysis tutorial. |
| Starter Panel Functions | Access to basic functions from the starter panel |
| Copyright holder | Cytoscape Consortium |
| How to use | By downloading cytoscape, you agree that you have read the license agreement that follows and agree to its terms. If you don't agree, do not download cytoscape |
For more in-depth information, visit the official Cytoscape website: https://cytoscape.org/
The power of Cytoscape lies not just in its core functionality but also in its extensibility. A vast ecosystem of apps expands its capabilities, catering to a wide array of problem domains. Whether delving into the intricacies of biological pathways, mapping social networks, or exploring the semantic web, Cytoscape offers tailored solutions through its app marketplace. Users can access basic functions through the starter panel when they start Cytoscape. The introduction to cytoscape and network biology is accessible. Various tutorials are also accessible like basic expression analysis tutorial and quick tour of cytoscape.
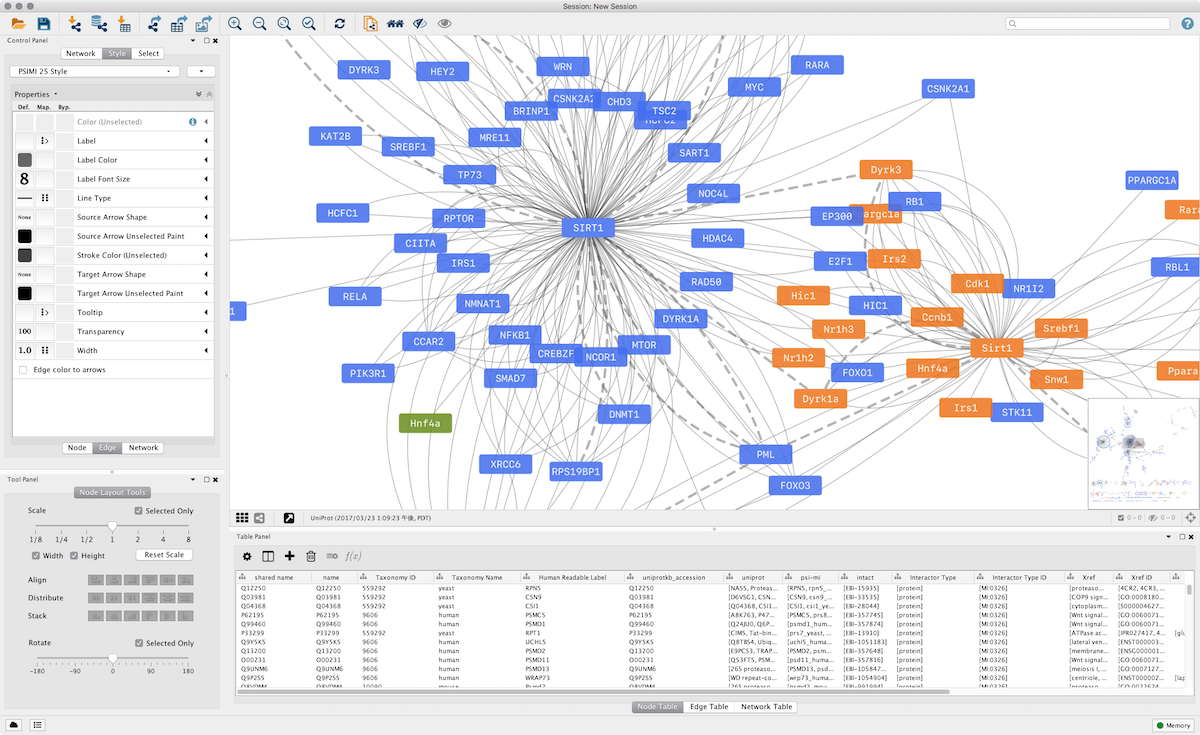
The user interface of Cytoscape is designed to be intuitive, providing a user-friendly experience for both novice and experienced users. A "Quick Tour" chapter in the manual offers a comprehensive overview of the basic layout and mechanics, guiding users through the essential steps of network creation, visualization, and analysis. For those who prefer a hands-on approach, the "Basic Expression Analysis Tutorial" provides a practical learning experience, allowing users to engage with the software and explore its functionalities directly.
Creating networks within Cytoscape is a versatile process, accommodating various data sources and user preferences. Users have multiple options when starting their projects: They can import data from public databases, which streamlines the process of accessing pre-existing datasets. Alternatively, users can create an empty network and manually add nodes and edges, providing complete control over the network's structure and composition. This flexibility ensures that Cytoscape can adapt to diverse research needs and project scopes.
The Cytoscape user manual is an invaluable resource for understanding and utilizing the software's full potential. It offers detailed explanations of the features, functionalities, and underlying principles of network analysis within Cytoscape. Both the Cytoscape 3.10.3 and Cytoscape 3.9.1 user manuals, which provide comprehensive documentation and guidance, are copyrighted by the Cytoscape consortium and are made available under the same GPL license as Cytoscape itself. This commitment to open-source principles ensures that the documentation is accessible and can be freely shared and adapted to meet user needs.
The platform's design emphasizes user accessibility and platform compatibility. Cytoscape is a Java application, verified to run seamlessly on Linux, Windows, and macOS platforms. While officially supported on these operating systems, its flexibility extends further. Although not officially supported, Cytoscape may also operate on other Unix platforms, such as Solaris or FreeBSD, provided that Java version 8 is available for that platform. This broad platform compatibility enhances its accessibility and utility for researchers and analysts across diverse computing environments.
Cytoscape's journey began in 2002 at the Institute of Systems Biology in Seattle. Since its inception, it has evolved and matured into a robust, widely-used platform. Today, development is driven by an international consortium of open-source developers, fostering a collaborative and dynamic environment that promotes innovation and ensures the platform's continued relevance. This collaborative approach allows for rapid updates and adaptation to changing research needs and technological advancements, strengthening Cytoscape's position as a leading tool in network analysis and visualization.
The open-source nature of Cytoscape underscores its commitment to collaboration and knowledge sharing. The platform and its documentation are freely available, enabling researchers and analysts worldwide to access and leverage its capabilities. This commitment to open-source principles fosters transparency, facilitates community contributions, and ensures that Cytoscape remains a dynamic and evolving resource for understanding complex networks. The GPL license that governs Cytoscape and its documentation guarantees the freedoms to use, modify, and distribute the software, promoting open collaboration and innovation within the scientific community.
The software is applicable to any system of molecular components and interactions, cytoscape is most powerful for handling complex networks with many interactions and attributes. It provides an excellent environment to analyze complex systems.
Before downloading Cytoscape, users are prompted to review and agree to the terms of the license agreement. This agreement ensures that users understand their rights and responsibilities when utilizing the software. By downloading the platform, users acknowledge that they have read and accepted the license, guaranteeing compliance with the terms of use and promoting ethical software practices. It is important to note that if users do not agree with the terms, they should refrain from downloading or using Cytoscape.
Cytoscape 3.0, represents a significant milestone in the platform's evolution. This version introduced a completely new architecture, which provides a modern and efficient foundation for future development. Moreover, it features a developer API, empowering developers to create custom apps and extensions tailored to their specific needs. This API promotes customization and expands the platform's capabilities, creating a dynamic ecosystem for innovation. Cytoscape 3.0 also incorporates a refined set of user controls, enhancing the user experience and simplifying navigation.
The open-source nature of Cytoscape is a cornerstone of its success. The platform's availability under a GPL license allows for its use, modification, and redistribution, fostering a community-driven approach to development and innovation. The core features and extensive app ecosystem are available to users without any cost. The open-source model encourages collaboration, transparency, and community contributions, fostering a vibrant ecosystem of developers and users who contribute to the platform's evolution and improvement.
In essence, Cytoscape stands as a testament to the power of open-source software in advancing scientific research and analytical capabilities. By providing a versatile and adaptable platform for visualizing and analyzing complex networks, Cytoscape empowers researchers and analysts across various disciplines to uncover hidden relationships and gain a deeper understanding of the world around them. With its intuitive user interface, robust features, extensive app ecosystem, and strong community support, Cytoscape remains a leading tool for exploring the intricacies of complex networks, supporting innovation and collaboration in the field of data analysis.